Clustering Techniques
Clustering
Cluster Analysis("data segmentation") is an exploratory method for identifying homogenous groups ("clusters") of records
- Similar records should belong to the same cluster
- Dissimilar records should belong to different clusters
- In Clustering there are two types of Clusters they are:
- Hierarchical Clustering
- Non-Hierarchical Clustering
Hierarchical Clustering Alogorithm:
- Hierarchical methods-agglomeratives: Begin with n clusters; sequentially merge similar clusters until 1 cluster is left. Useful when goal is to arrange the clusters into a natural hierarchy. Requires specifying distance measure
Clustering
Cluster Analysis(“data segmentation”) is an exploratory method for identifying homogeneous groups
(“clusters”) of records
- Similar records should belongs to same cluster
- Dissimilar records should belongs to different clusters
- In clustering there are two types of clusters they are:
- Hierarchical clustering
- Non-Hierarchical clustering
Hierarchical clustering Algorithm:
- Hierarchical methods - Agglomerative
Begin with n clusters;sequentially merge similar clusters until 1 cluster is left.
Useful when the goal is to arrange the clusters into natural hierarchy.
Requires specifying distance measures
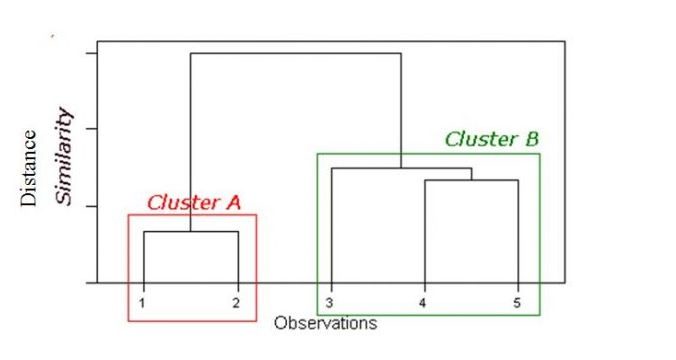
Dendrogram:
Tree like diagram that summarise the clustering process
Similar records join by links
Record location determined by similarity to other records
- Start with n clusters
( 1 record in each cluster)
- Step 1: Two closest records merged into one cluster
- At every step, pairs of records/clusters with smallest distance are merged.
- Two records are merged,
- or single record added to the existing cluster,
- or two existing clusters are combined
- requires a definition of distance.
Pairwise distance between records:
dij = distance between records i and j
Distance requirements : Non-negative(dij>0) dii =0
Symmetry(dij = dji)
Triangle inequality (dij+djk>=djk)
Distance for binary data
- Binary Euclidean Distance : (b+c) / (a+b+c+d)
- Simple matching coefficient : (a+d) / (a+b+c+d)
- Jaquard’s coefficient : d / (b+c+d)
for>2 categories,distance=0 only if both items have same category.Otherwise=1
Distances for mixed(Numerical+categorical)Data
- Simple:Standardized numerical variables to [0,1],the use euclidean distance for all
- Gower’s general dissimilarity coefficient
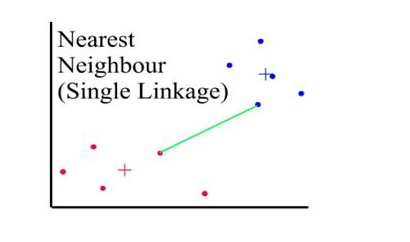
Distances between clusters: ;’Single linkage’(‘nearest neighbour’)
- Distance between 2 clusters = minimum distance between members of the two clusters
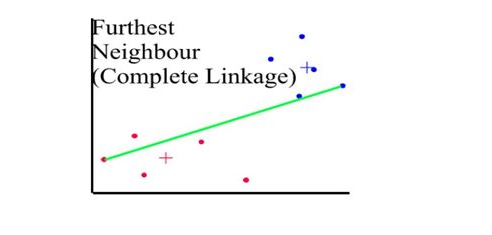
Distances between clusters: ;’complete linkage’(‘farthest neighbour’)
- Distance between 2 clusters = Greatest distance between members of the two clusters
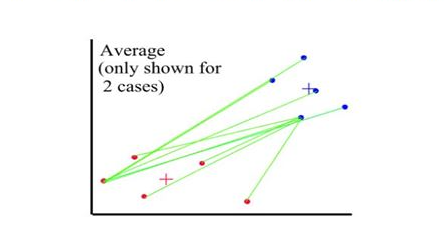
Distances between clusters: ; ‘Average linkage’
- Distance between 2 clusters = Average of all distances between members of the two clusters
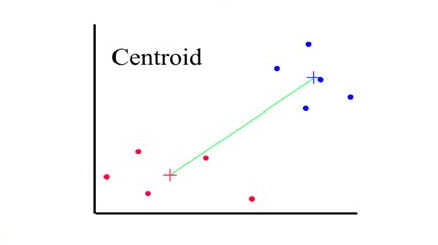
Distances between clusters: ; ‘Centroid linkage’
- Distance between 2 clusters = Distance between centroids(centers)
Hierarchical
The good
- Finds “natural” grouping - no need to specify number of clusters
- Dendrogram: transparency of process, good for presentation
The Bad
- Require computation and storage of n/n distance matrix
- Algorithm makes only one pass through the data.records that are incorrectly allocated early on cannot be reallocated subsequently
- Low stability : recording data or dropping a few records can leads to different solution
- single complete linkage robust to distance metrics as long as the relative ordering is kept .
- Average linkage is Not.
- Most distances sensitive to outliers.
Non-Hierarchical methods:
K-means clustering
- pre-specified number of clusters ,assign records to each of the clusters.
- Requires specifying # clusters
- Computationally cheap
- Predetermined number(k) of non-overlapping clusters
- Clusters are homogenous yet dissimilar to other clusters
- Need measures of within-cluster similarity(homogeneity) and between-cluster similarity
- No hierarchy(no dendrogram)!End-product is final cluster memberships
- Useful for large datasets
Algorithm minimizes within-cluster variance(heterogeneity)
1.For a user-specified value of k,partition dataset into k initial clusters
2.For each record,assign it to cluster with closest centroid
3.Re-calculate centroids for the “losing” and “receiving” clusters.Can be done
- After reassignment of each record,or
- After one complete pass through all records(cheaper)
4.Repeat step 2-3 until no more assignment is necessary
Initial partition into k clusters
Initial partitions can be obtained by either
- User-specified initial partitions,or
- User-specified initial centroids,or
- Random partitions
Stability : run algorithm with different initial partitions
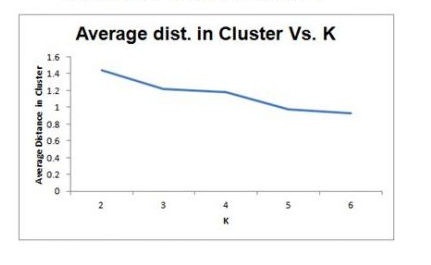
Selecting K
- Re-run algorithm for different values of k
- Trade off: simplicity (interpretation) vs.adequacy ( within-cluster homogeneity)
- Plot within-cluster variability as a function of k
K-Means
The Good
- Computationally fast for large datasets
- Useful when certain k needed
The Bad
- Can take long to terminate
- Final solution not guaranteed to be “globally optimal”
- Different initial partitions can lead to different solutions
- Must re-run the algorithm for different values of K No dendrogram